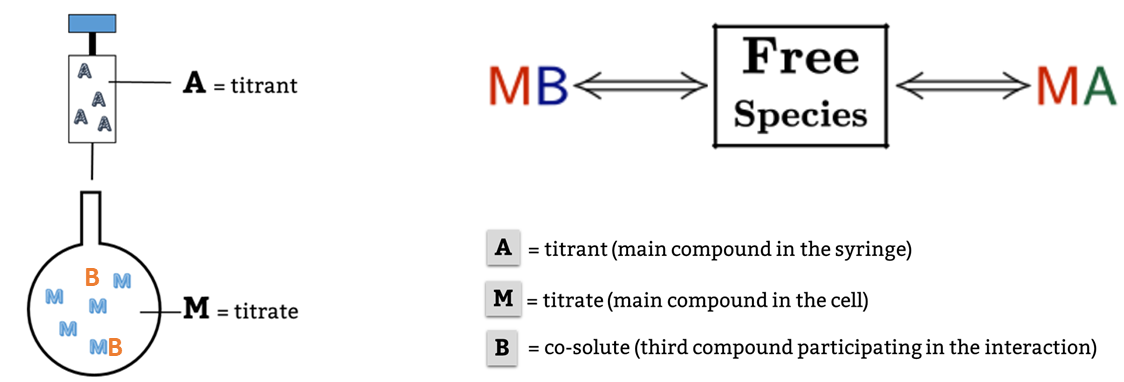
AFFINImeter is already well known in ITC binding data analysis for providing the possibility to use tailored binding models created by the user. The models are generated with the tool “model builder” that includes a letter code “M-A-B” to describe titrate (M), titrant (A) and if necessary, the presence of a third species (B) (Figure 1).
Fig.1 Example of a competitive binding model created in AFFINImeter where the titrant in syringe “A” binds to the titrate in cell “M” to form a 1:1 complex “MA” and a second ligand “B” mixed in the cell with “M” forms the complex “MB” and thus competes with “A”.
Following the same approach, the binding models available for the software AFFINImeter for Nuclear Magnetic Resonance are generated with the model builder and based on the “M–A–B” code. But there are significant differences between ITC and NMR data analysis when the time comes to select a binding model from AFFINImeter, which have an origin in the inherent characteristics of each technique and in the different experimental design. In chemical shift perturbation (CSP) NMR titration experiments, the observed parameter used to monitor the progress of the binding event is the chemical shift of titrate resonance signals. Hence, the models used for NMR data analysis require the presence of compound “M” (titrate) as it is the species from which changes associated with the binding process are monitored. Conversely, in ITC the observed parameter is the heat change upon interaction and this parameter is not necessarily linked to a particular species “M”, “A” or “B”.
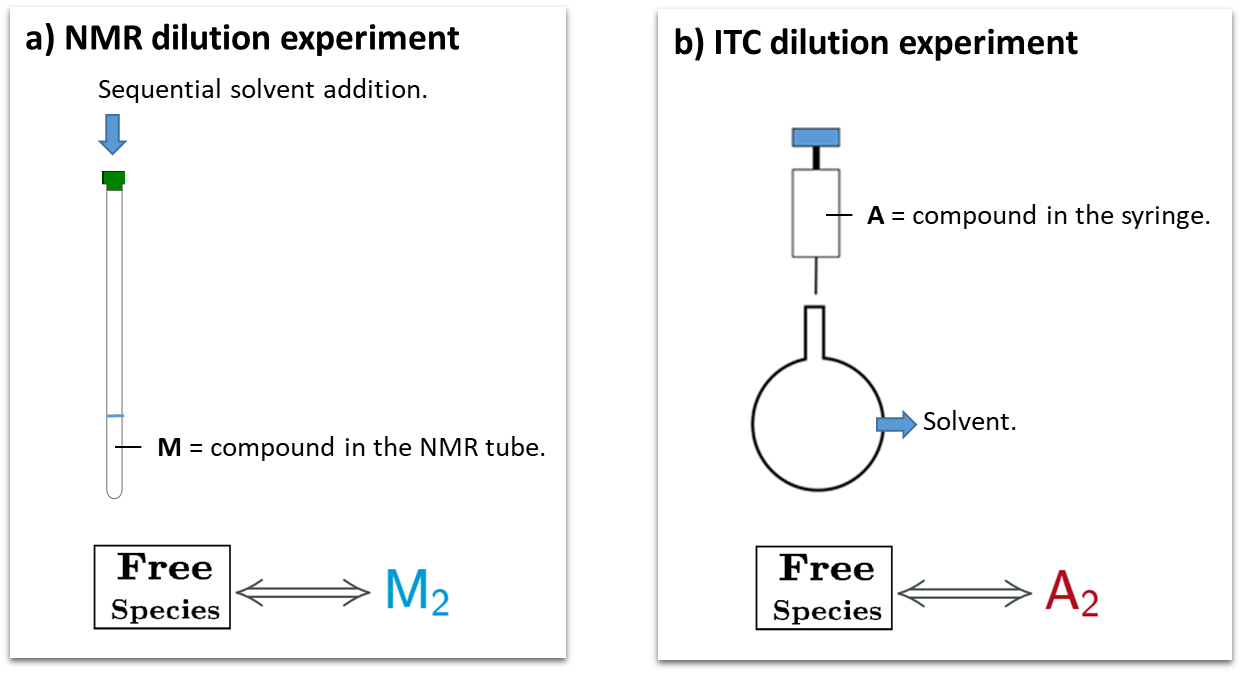
An illustrative example is the evaluation of a monomer-dimer self-association process using NMR or ITC. In NMR, the standard experimental setup would consist in the incremental dilution of the compound sample at high concentration in the NMR tube, to monitor dimer dissociation (Figure 2a). In ITC the standard experimental setup would consist in a titration of the compound sample at high concentration in the syringe (species “A” according to the AFFINImeter code) into the calorimetric cell filled up with solvent (Figure 2b).
Fig.2 Representation of experimental setup for a) NMR dilution experiment and b) ITC dilution experiments. The corresponding schemes of AFFINImeter binding models for data analysis are shown.
Would you like to know more about AFFINImeter for Nuclear Magnetic Resonance? Press the button below: