Isothermal titration Calorimetry experiments of a ligand binding to a macromolecule with multiple independent sites
A successful Isothermal Titration Calorimetry (ITC) experiment requires the acquisition of high quality experimental data together with a careful analysis. Choosing the right binding model to fit the ITC isotherm is critical in order to get the true thermodynamic profile of the interaction. Often, the main limitation to achieve good results arises when the evaluation software lacks of the mathematical model that best describes our binding experiment. A good example is the case of a ligand binding to a macromolecule with multiple independent sites, i.e ligand – DNA interactions (1). Until now the readily available mathematical models to fit such experiments was limited to one or two sets of “n” independent identical sites; frequently, these models offer a poor description of the interaction due to the inherent higher complexity of the system, where many distinct binding equilibria coexist.
Multiple Sets of Independent Sites
AFFINImeter ITC offers an unlimited number of user-defined binding models. Particularly, it counts with a feature to easily design models based on multiple independent binding sites. Here, a model with a number of sets of independent sites can be created with no limitation in the number of sets or sites. Noteworthy, the number of sites in each set can be considered as a fitting parameter throughout the data analysis. As an illustration, the following figure shows the reaction parameters of a model generated with AFFINImeter that describes a ligand binding to a receptor having 3 sets of sites, each set having an unknown number of sites. Fitting the experimental data to such model yields the microscopic association constant (K) and the change in enthalpy (ΔH) of the ligand binding to each site type, and the number of sites in each set (n).
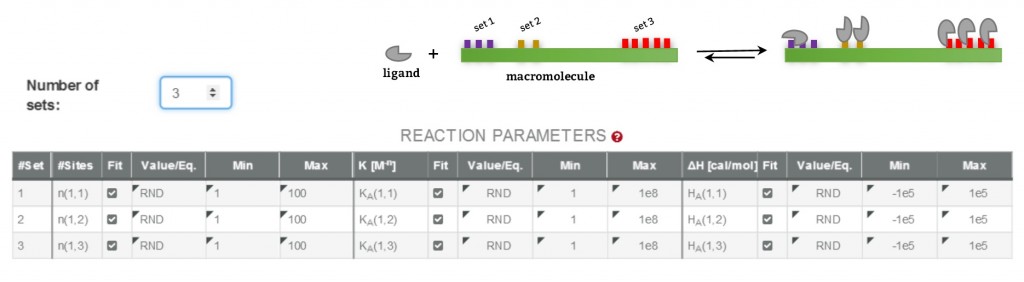
These binding models, described by numerous variable parameters, may end up in an over-parameterized fitting function. Thus, the best strategy to achieve a robust and consistent analysis involves the global fitting of several ITC curves acquired under different experimental conditions. In this sense, AFFINImeter also supports global fitting of multiple isotherms wherein parameter linkage between curves is used to decrease the relative number of estimated parameters per experiment.
References:
(1) Methods 2007, 42, 162–172.